Phage display technologies
Phage display technologies allow the identification of peptide ligands for a given target molecule out of a huge library of different peptides expressed on the surface of bacteriophages.f Bakteriophagen präsentiert.
Phage display technologies allow the identification of peptide ligands for a given target molecule out of a huge library of different peptides expressed on the surface of bacteriophages. Presentation of the peptide library on the surface of bacteriophages ("phage display") as a fusion of peptide and a phage coat protein allows the physical link between the presented peptide and the DNA sequence coding for its amino acid sequence. Diversity of the peptides/proteins can be introduced by combinatorial mutagenesis of the fusion gene. Extremely large numbers of different peptides can be constructed, replicated, selected and amplified in a process called "biopanning". The libraries are incubated with a target molecule either as an immobilized target or prior to capture of the target-phage complex on a solid support. Similar to affinity chromatography, non-interacting peptides and proteins are washed off and the interacting molecules are subsequently eluted. The interacting phage displayed peptides or proteins can be amplified by bacterial infection to increase their copy number. The selection/amplification process can be repeated to further enrich those library members with relatively higher affinity to the target. The result is a final peptide population that is dominated by sequences that bind the target with highest affinity.
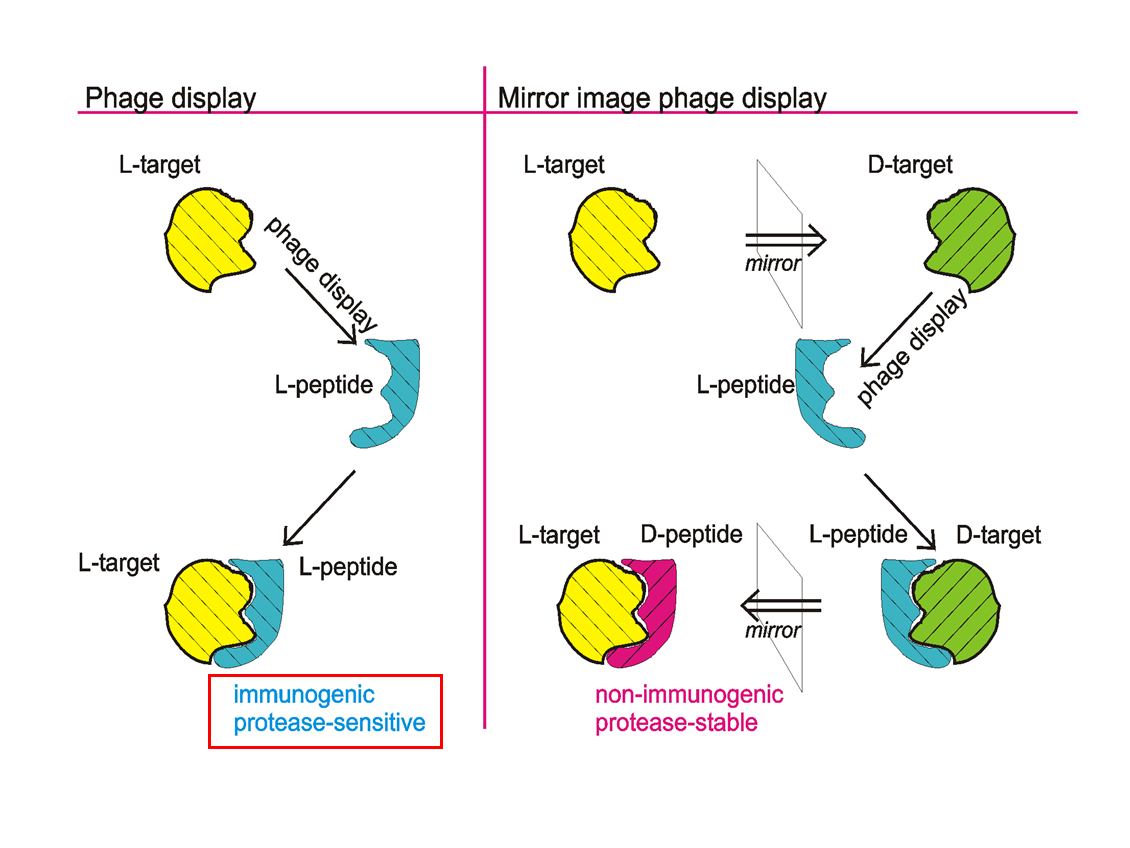
IIn therapeutic or diagnostic in vivo approaches, D-enantiomeric peptides have several advantages over L-enantiomeric peptides. Most importantly, they are resistant to proteases, which can dramatically increase serum and saliva half-life. A very elegant and attractive way to obtain D-peptides that bind to specific targets is mirror image phage display. During a common phage display approach, as invented in 1990, a L-peptide library is presented on the surface of phages and selected for those L-peptides (e.g. by biopanning) that bind to a given target protein. In mirror image phage display, the selection is carried out against the mirror image of the original target, which is in the case of proteins a protein with the same amino acid sequence but fully composed of D-enantiomeric amino acids (see figure). Mirror image phage display uses the full advantages of phage display, allowing the relatively easy and straight forward selection of ligands out of a huge biologically encoded library that consists of up to 1013 different peptide or protein variants, thereby also delivering a rich source of structural diversity.